Calibration¶
Contents
Félix-Antoine Fortin, François-Michel De Rainville, Marc-André Gardner, Marc Parizeau and Christian Gagné, “DEAP: Evolutionary Algorithms Made Easy”, Journal of Machine Learning Research, vol. 13, pp. 2171-2175
Note
You can install it with: Pip install deap
Calibration method¶
Calibration is using an evolutionary computation framework in Python called DEAP (Fortin et al., 2012). We used the implemented evolutionary algorithm NSGA-II (Deb et al., 2002) for single objective optimization. As objective function we used the modified version of the Kling-Gupta Efficiency (Kling et al., 2012), 2012), with r as the correlation coefficient between simulated and observed discharge (dimensionless), β as the bias ratio (dimensionless) and γ as the variability ratio.
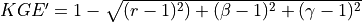
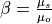 and
and 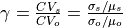
Where CV is the coefficient of variation, μ is the mean streamflow [m3 s−1] and σ is the standard deviation of the streamflow [m3 s−1]. KGE’, r, β and γ have their optimum at unity. The KGE’ measures the Euclidean distance from the ideal point (unity) of the Pareto front and can therefore provide an optimal solution that is simultaneously good for bias, flow variability, and correlation. For a discussion of the KGE objective function and its advantages over the often used Nash–Sutcliffe Efficiency (NSE) or the related mean squared error, see (Gupta et al., 2009). The calibration uses a general population size (µ) of 256, a recombination pool size (λ) of 32.The number of generations was set to 30, which we found was sufficient to achieve convergence for stations
Suggested Calibration parameters¶
Calibration tool structure¶
calibration
│- readme.txt
│- readme.txt
│
└--observed_data
│ └- lobith2006.cvs, ...
│
└--templates
│ └-- runpy.bat, runpy.sh
│ └-- settings.ini
How it works¶
The calibration tool builds up a single-objective obtimization framework using the Python libray DEAP For each run it triggers the run of the hydrological model:
using a template of the settings file
replacing the output folder in this template file
replace placeholders with the values of calibration parameters, the limit of the parameter range is given in the file: ParamRanges.csv
After each run the model run is compared to observed values (e.g. observed_data/lobith2006.csv)
After the calibration, statistics and the best run is printed output
What is needed¶
runpy.bat: the path to cwatm.py have to be set correctly (for linux a .sh file has to be created)
The actual version of a cwatm settings file has to modified:
replacing the output folder with the placeholder: %run_rand_id
28#-------------------------------------------------------
29# CALIBARTION PARAMETERS
30#-------------------------------------------------------
31[CALIBRATION]
32
33# These are parameter which are used for calibration
34# could be any parameter, but for an easier overview, tehey are collected here
35# in the calibration template a placeholder (e.g. %arnoBeta) instead of value
36
37OUT_Dir = %run_rand_id
putting the output variables in e.g. OUT_TSS_Daily = discharge or monthly average discharge OUT_TSS_MonthAvg = discharge
38OUT_TSS_Daily = discharge
39OUT_TSS_MonthAvg = discharge
delete all the output variables in the template (mostly at the end of the file)
replacing calibration parameter values with a placeholder: e.g. %SnowMelt
42# Snow SnowMeltCoef = 0.004
43SnowMeltCoef = %SnowMelt
44# Cropf factor correction
45crop_correct = %crop
46#Soil
47soildepth_factor = %soildepthF
48#Soil preferentialFlowConstant = 4.0, arnoBeta_factor = 1.0
49preferentialFlowConstant = %pref
50arnoBeta_add = %arnoB
51# interflow part of recharge factor = 1.0
52factor_interflow = %interF
53# groundwater recessionCoeff_factor = 1.0
54recessionCoeff_factor = %reces
55# runoff concentration factor runoffConc_factor = 1.0
56runoffConc_factor = %runoff
57#Routing manningsN factor [0.1 - 10.0] default 1.0
58manningsN = %CCM
59# reservoir normal storage limit (fraction of total storage, [-]) [0.15 - 0.85] default 0.5
60normalStorageLimit = %normalStorageLimit
61# lake parameter - factor to alpha: parameter of of channel width and weir coefficient [0.33 - 3.] dafault 1.
62lakeAFactor = %lakeAFactor
63# lake wind factor - factor to evaporation from lake [0.8 - 2.] dafault 1.
64lakeEvaFactor = %lakeEvaFactor
ParameterName,MinValue,MaxValue
SnowMelt,0.001,0.007
crop,0.8,3.0
soildepthF, 0.8,1.8
pref,0.5,8
arnoB,0.01,1.0
interF, 0.33,3.0
reces,0.1,10
runoff,0.1,5
CCM,0.1,10.0
normalStorageLimit,0.15,0.85
lakeAFactor,0.333,3.0
lakeEvaFactor,0.5,3.0
No,1,100
1#-------------------------------------------------------
2[TIME-RELATED_CONSTANTS]
3#-------------------------------------------------------
4
5# StepStart has to be a date e.g. 01/06/1990
6# SpinUp or StepEnd either date or numbers
7# SpinUp: from this date output is generated (up to this day: warm up)
8
9StepStart = 1/1/1990
10SpinUp = 1/1/1995
11StepEnd = 31/12/2010
[DEFAULT]
Root = /c/watmodel/CWATM
RootPC = C:/watmodel/CWATM
Rootbasin = calibration_rhine
ForcingStart = 1/1/2000
ForcingEnd = 31/12/2010
timeperiod = daily
[ObservedData]
Qtss = observed_data/lobith.csv
Column = lobith
Header = River: Rhine station: Lobith
[Validate]
Qtss = observed_data/lobith_val.csv
ValStart = 1/1/1990
ValEnd = 31/12/1999
[Path]
Templates = templates
SubCatchmentPath = catchments
ParamRanges = ParamRanges.csv
[Templates]
ModelSettings = settings.ini
RunModel = runpy.sh
[Option]
firstrun = False
para_first = [0.0022, 1.72, 1.24, 7.07, 0.55, 1.92, 2.81, 0.74,1.34,0.35,2.04,1.0, 1.]
# Snowmelt, crop KC, soil depth,pref. flow, arno beta, interflow factor, groundwater recession,
# runoff conc., routing, manning factor, normalStorageLimit, lakeAFactor,lakeEvaFactor,No of run
bestrun = True
[DEAP]
maximize = True
use_multiprocessing = 1
ngen = 30
mu = 256
lambda_ = 32
Recommendations¶
146[INITITIAL CONDITIONS]
147#-------------------------------------------------------
148
149# for a warm start initial variables a loaded
150# e.g for a start on 01/01/2010 load variable from 31/12/2009
151load_initial = False
152initLoad = $(FILE_PATHS:PathRoot)/init/Rhine_19891231.nc
153
154# saving variables from this run, to initiate a warm start next run
155# StepInit = saving date, can be more than one: 10/01/1973 20/01/1973
156save_initial = False
157initSave = $(FILE_PATHS:PathRoot)/init/Rhine
158StepInit = 31/12/1989 31/12/2010
Running calibration¶
Look into the settings file of the calibration folder.
look into runCalibration.bat. If Python is in your computer path everything should be ok, otherwise put in the path to Python
look into templates/runpy.bat. Put the path to Python in if necessary
look into templates/settings.ini. Put the paths in a right way that it fits your computer:
[FILE_PATHS] #------------------------------------------------------- PathRoot = P:/watmodel/CWatM/calibration_tutorial PathOut = $(PathRoot)/output PathMaps = $(PathRoot)/CWatM_data/cwatm_input PathMeteo = $(PathRoot)/climate
in observed_data/yukon2001.cvs you find the observed data:
- make sure the name in the header is the same as in [ObservedData] Column - make sure that there are enough data in (from ForcingStart to ForcingEnd)
make sure the folder catchments is empty! Before each try this folder has to be empty
Run runCalibration.bat¶
go for testing (see below)
go for testing again (see below)
Change use_multiprocessing =1 in settings.txt
Run runCalibration.bat and after some time something should appear in your window
For testing¶
Change use_multiprocessing = 0 in settings.txt
Delete catchments but keep the empty folder
Run runCalibration.bat and wait till catchment/00_001 gets filled, then interrupt
Change to catchments/00_001
Run runpy00_001.bat
See what errors come up and change settings-Run00_001.ini
Change template/setting.ini in the same way
Do this again and again till no error
Running it on your computer¶
It will be really slow on Windows using data on the server – next step run it on your PC
copy the whole folder P:watmodelCWatMcalibration_tutorial to your PC (only 15 GB)
(but maybe you have already parts of it on your computer – like the big climate input files)
Make it work on your computer:
Changing file paths in templates/settings.ini, setting.txt Changing the path for Python in runCalibration.bat and templates/runpy.bat
Preparation for another catchment¶
Preparing the observed dataset – discharge¶
Calibration works by comparing simulated discharge with observed discharge using an objective function: Here we use the Kling-Gupta Efficiency but we can also use Nash-Sutcliffe Efficiency . Please find some more information on the objective function an on the evolutionary computation framework used for calibration on: https://cwatm.github.io/calibration.html
The observed values can be stored as daily values or monthly values
The observed values should be at least cover 5 years (best is 10-15 years)
The observed discharge has to be stored as textfile in:
./observed_data/nameofstation.cvs And has to look like this: date,yukon_pilot_station 2001-04-01,1302.6 2001-04-02,1302.6 2001-04-03,1302.6 2001-04-04,1302.6 … … 2013-12-31,2647.6
Or:
date ,zhutuo 2002-01-01,3229.0 2002-02-01,2979.2 2002-03-01,3229.0
Format:
Date format like this year-month-day [yyyy-mm-dd]
Separated by a comma
Discharge in [m3/s]
If a value is missing, that is not a problem (as long as the time series is long enough):
it should look like this: (no value after the comma) 2002-01-12,
For each day (or month) a line
Settings.txt
In the settings file, the lines:
[ObservedData]
Qtss = observed_data/zhutuo_2002month.csv
Column = zhutuo
Header = River: Yangtze station: Zhutuo
Should correspond to the name and header in the observed discharge.cvs
The lines:
ForcingStart = 1/1/2002
ForcingEnd = 31/12/2013
Should correspond to the number of lines in the observed discharge.cvs
Creating an initial netcdf file for warm start¶
It is best to have a long warm-up phase, especially for groundwater: See also: https://cwatm.github.io/setup.html#initialisation
You can run CWatM for a couple of years (20 years or more) and store the last days storage values in a file. This file can be read in to enable a ‘warm” start
change use_multiprocessing = 0 in settings.txt
Delete catchments but keep the empty folder
Run runCalibration.bat and wait till catchment/00_001 gets filled, then interrupt
Change to catchments/00_001
Open the settings-Run_001.init
Change load_initial = True to load_initial = False
save_initial = True
initSave = $(FILE_PATHS:PathRoot)/CWatM_init/testx
StepInit = 31/01/1996 (change it to a date 1 month after your StepStart)
Run runpy00_001.bat
There should be a file ./CWatM_init/testx_19960131.nc
Change to: load_initial = True
initLoad = $(FILE_PATHS:PathRoot)/CWatM_init/testx_19960131.nc
Run runpy00_001.bat
If it works, then it used the initial file you generated before (that was just a test)
Now change to:
StepStart = 1/1/1961
StepEnd = 31/12/2013
load_initial = False
save_initial = True
initSave = $(FILE_PATHS:PathRoot)/CWatM_init/station_name
StepInit = 31/12/2013
Run runpy00_001.bat
This should have generated a file ./CWatM_init/station_name_20131231.nc
And again:
StepStart = 1/1/1961 (some 20 years or longer)
StepEnd = 31/12/1995 (a day before your normal running day)
load_initial = True
initLoad = $(FILE_PATHS:PathRoot)/CWatM_init/station_name_20131231.nc
save_initial = True
initSave = $(FILE_PATHS:PathRoot)/CWatM_init/station_name
StepInit = 31/12/1995 (a day before your running day)
Run runpy00_001.bat
This should have generated a file ./CWatM_init/station_name_19951231.nc
And last part:
Change StepStart and StepEnd back to original values
load_initial = True
initLoad = $(FILE_PATHS:PathRoot)/CWatM_init/station_name_19951231.nc
save_initial = False
Run runpy00_001.bat
If it works, do the same in the ./template/settings.ini
Note
You have now a “warm” start for every calibration run
Cutting out a catchment as mask map¶
See the .doc file in P:watmodelCWatMcalibration_tutorialcalibrationtoolscut_catchmentFor a description:
Requirements: PCRASTER:
We do not need the Python version, I think downloading, extracting and setting of the paths in P:watmodelCWatMcalibration_tutorialcalibrationtoolscut_catchmentcatchconfig_win.ini Creating the 2 potential evaporation files in advance
Potential evaporation is calculated with the Penman-Monteith equation in CWatM, but it is not part of the calibration—there is no change in pot. Evaporation. To make calibration computation faster, the results of pot evaporation could be stored and reused each time.
For the 30min, this is already done as a global map set, but for the 5min, these files become too big. So they have to be produced for each basin separately
Same preparation as for Creating an initial netcdf file for warm start see above There should be a folder catchments00_001 with a working run for 001.
Open the settings-Run_001.init
Change:
[Option] calc_evaporation = True
[TIME-RELATED_CONSTANTS] SpinUp = None
[EVAPORATION]
OUT_Dir = $(FILE_PATHS:PathOut)
OUT_MAP_Daily = ETRef, EWRef
Run runpy00_001.bat
There should be a file ETRef.nc and EWRef in the output directory.
Rename the files e.g. ETRef.nc to ETRef_yangtze.nc, EWRef.nc to EWRef_yangtze.nc and copy it to PathMeteo (or somewhere else, you have to put the path in)
Open the settings-Run_001.init
Change:
[Option] calc_evaporation = False
[TIME-RELATED_CONSTANTS] SpinUp = -> to the time it was before
[Meteo]
daily reference evaporation (free water)
E0Maps = $(FILE_PATHS:PathMeteo)/EWRef_yangtze
daily reference evapotranspiration (crop)
ETMaps = $(FILE_PATHS:PathMeteo)/ETRef_yangtze
[EVAPORATION]
OUT_Dir = $(FILE_PATHS:PathOut) !!! outcomment this again - important
OUT_MAP_Daily = ETRef, EWRef
Test it: Run runpy00_001.bat
And change the settings.ini in templates in the same way
Calibration of a downstream catchment¶
Calibration of a downstream catchment (upstream catchment is already calibrated) can be done using:
The catchment area of the downstream catchment minus the upstream catchment
The missing discharge from the upstream catchment is replaced by an inflow file
Cut the mask map, so that the upstream catchment is NOT in the mask map anymore
Detect the point(s) downstream of the inflow points
Run the best calibration scenario(s) of the upstream catchments again to produce long timeserie(s) of the outlet(s) point
Create an inflow file from the long timeseries of outlet(s)
Create a downstream calibration settings (directories, templates etc.)
Test the catchment!
Change the settings file of the downstream calibration so that it includes the inflow from upstream
Test it! 7. Create initial file for warm start
Cutting the mask map¶
Assuming you have a mask map of the whole catchment (e.g. Yangtze.map and the station points (here Zhutuo 105.75 28.75 and Yichang 111.25 30.75 1. Creating catchment for Zhutuo: catchment 105.75 28.75 ldd_yangtze.map zhu1.map 2. Creating catchment for Yichang: catchment 111.25 30.75 ldd_yangtze.map yi1.map 3. Creating Yichang without Zhutuo:
pcrcalc a2.map = cover(scalar(zhu1.map)*2,scalar(yi1.map))
pcrcalc yichang.map = boolean(if(a2.map eq 1,a2.map))
Result is a maskmap: Yichang.map
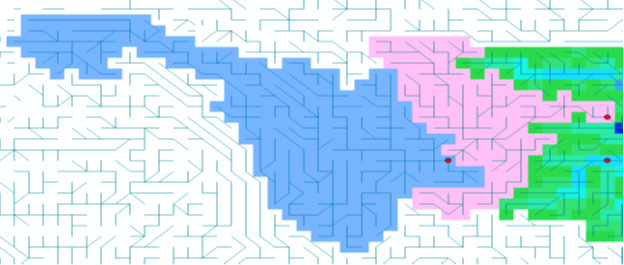
Figure 1: Upstream catchment (blue) and downstream catchment (red)
Detecting the downstream point¶
The inflow point of the new catchment has to be in the new mask and preferable one grid cell in flow direction below the upstream station e.g. 1 gridcell North East of Zhutuo (see purple circle in fig. 2)
The inflow point has the lon/lat 106.25 29.25
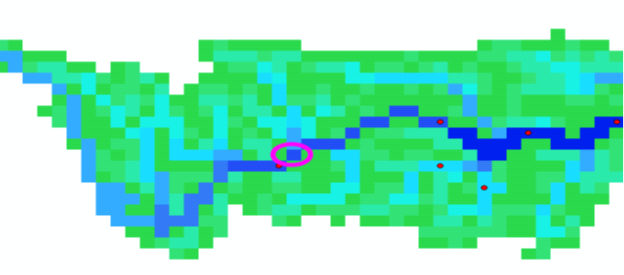
Figure 2: Downstream point
Run the best calibration scenario upstream¶
In order to get a long inflow time series for the inflow point (here: Zhutuo), you need to run the best scenario of the upstream catchment (here: 31_best)
Change into the folder ../catchments/best
Change settings file from:
StepStart = 1/1/1996 SpinUp = 1/1/2002 StepEnd = 31/12/2013
To:
StepStart = 1/1/1990 SpinUp = 1/1/1996 StepEnd = 31/12/2013
Results is a time series from 1/1/1990 – 31/12/2013 in: discharge_daily.tss
Create an inflow file from the long timeseries of outlet(s)¶
Create a folder ../inflow
Copy the ../catchments/31_best/discharge_daily.tss to ../inflow/zhutuo.tss
Create a downstream calibration settings (directories, templates etc.)¶
Create downstream calibration settings as before
Copy everything from upstream catchment (e.g. zhutuo) but not catchments
Create empty catchments folder
Create a observed discharge file in observed
Change settings.txt accordingly
Change settings.ini accordingly
Test the catchment setting!
But do not create an initial run yet!
Change the settings file¶
Change the settings file of the downstream calibration so that it includes the inflow from upstream Change the part of the settings.ini:
[Option]
inflow = True
[INFLOW]
#-------------------------------------------------------
# if option inflow = true
# the inflow from outside is added as inflowpoints
In_Dir = $(FILE_PATHS:PathRoot)/calibration/calibration_yichang/inflow
# nominal map with locations of (measured)inflow hydrographs [cu m / s]
InflowPoints = 106.25 29.25
InLocal = True
.
# if InflowPoints is a map, this flag is to identify if it is global (False) or local (True)
# observed or simulated input hydrographs as time series [cu m / s]
# Note: that identifiers in time series have to correspond to InflowPoints
# can be several time series in one file or different files e.g. main.tss mosel.tss
QInTS = zhutuo.tss
Test it!
Generate initial file for warm start Use initial file for calibration
Joining best sub-basin results to calibration maps¶
You need all runs done for all sub-basins
A region map
For each subbasin a unique number e.g. Zambezi basin

Figure 3 Sub-basin map with a unique identifier for each subbasin
You need a working PCRaster installation
The settings file settings.txt has to be changed:
[DEFAULT] Root = P:/watmodel/CWatM/calibration/calibration_zambezi # root directory where all subbasin are in . [Catchments] catch = lukulu, katima, kafue, luangwa, kwando, tete # name of the subbasin, has to be the same as the folder name in root # the order has to be the same as in the region map . [region] regionmap = P:/watmodel/CWatM/calibration_tutorial/calibration/CreateCalibrationMaps/zambezi_regions.map # region map, the order has to be the same a [Catchment] . [Path] Templates = %(Root)s/templates SubCatchmentPath = %(Root)s/catchments ParamRanges = %(Root)s/Join/ParamRanges.csv . Result = P:/watmodel/CWatM/calibration_tutorial/calibration/CreateCalibrationMaps/results # here are the results . PCRHOME = C:\PCRaster\bin # Where is your PCraster installation?
Run Python CAL_5_PARAMETER_MAPS.py
References¶
Beck, H. E., A. I. J. M. van Dijk, A. de Roo, D. G. Miralles, T. R. McVicar, J. Schellekens and L. A. Bruijnzeel (2016). “Global-scale regionalization of hydrologic model parameters.” Water Resources Research 52(5): 3599-3622.
Deb, K., A. Pratap, S. Agarwal and T. Meyarivan (2002). “A fast and elitist multiobjective genetic algorithm: NSGA-II.” IEEE Transactions on Evolutionary Computation 6(2): 182-197.
Fortin, F. A., F. M. De Rainville, M. A. Gardner, M. Parizeau and C. Gagńe (2012). “DEAP: Evolutionary algorithms made easy.” Journal of Machine Learning Research 13: 2171-2175.
Greve, P., L. Gudmundsson, B. Orlowsky and S. I. Seneviratne (2016). “A two-parameter Budyko function to represent conditions under which evapotranspiration exceeds precipitation.” Hydrology and Earth System Sciences 20(6): 2195-2205.
Gupta, H. V., H. Kling, K. K. Yilmaz and G. F. Martinez (2009). “Decomposition of the mean squared error and NSE performance criteria: Implications for improving hydrological modelling.” Journal of Hydrology 377(1-2): 80-91.
Kling, H., M. Fuchs and M. Paulin (2012). “Runoff conditions in the upper Danube basin under an ensemble of climate change scenarios.” Journal of Hydrology 424-425: 264-277.
Samaniego, L., R. Kumar, S. Thober, O. Rakovec, M. Zink, N. Wanders, S. Eisner, H. Müller Schmied, E. Sutanudjaja, K. Warrach-Sagi and S. Attinger (2017). “Toward seamless hydrologic predictions across spatial scales.” Hydrology and Earth System Sciences 21(9): 4323-4346.